
The target for Hiro Clinic’s testing accuracy (probability of paternity inclusion) is
99.99999999999% or higher is our target.
If we assume a global population of 8 billion people, the probability of a mismatch would be only 0.008 individuals. This result is derived using a statistical measure called the likelihood ratio, and it is achievable through the use of STR analysis.
Relationship Between Likelihood Ratio and Probability of Paternity
| Likelihood Ratio | Probability of Paternity | Probability of Error |
|---|---|---|
| 10 | Over 90% | 1 in 10 people |
| 1,000 (3 zeros) | Over 99.9% (3 nines) | 1 in 1,000 people |
| 100,000 (5 zeros) | Over 99.999% (5 nines) | 1 in 100,000 people |
| 10,000,000 (7 zeros) | Over 99.99999% (7 nines) | 1 in 10,000,000 people |
| 1,000,000,000 (9 zeros) | Over 99.9999999% (9 nines) | 1 in 1,000,000,000 people |
| 100,000,000,000 (11 zeros) | Over 99.999999999% (11 nines) | 1 in 100,000,000,000 people |
| 10,000,000,000,000 (13 zeros) | Over 99.99999999999% (13 nines) | 1 in 10,000,000,000,000 people |
Because STRs (Short Tandem Repeats) have a wide range of numerical variations, they allow for more precise differentiation.
To achieve this level of accuracy using SNPs (Single Nucleotide Polymorphisms) alone, more than 5,000 SNPs would be required.
Modern Advancements in DNA Paternity Testing Technology
The ABO blood group system was discovered in 1900 and began to be used for paternity exclusion testing in the 1930s. Later, the inclusion of additional blood markers such as the Rh antigen, MNS antigens, and human leukocyte antigens (HLA) improved testing accuracy. However, these methods could not completely eliminate errors.
In 1977, the development of DNA sequencing through the Sanger method enabled direct analysis of DNA, achieving greater statistical accuracy in paternity testing compared to traditional techniques. Today, DNA testing is widely used not only for paternity testing but also in forensic investigations, prenatal testing, and many other applications.
Testing Process
If the test results are intended for use in legal proceedings, the samples from the parties involved are collected at the clinic by a physician. In all other cases, samples can be easily collected at home using a simple and user-friendly kit, and then sent back to the laboratory.
Upon receipt, the samples are registered in the laboratory database. The collected cells then undergo DNA extraction and purification from the cell nuclei. This process is performed using a fully automated, high-throughput system, achieving shorter processing times, lower costs, and higher accuracy compared to traditional DNA testing methods.
Additionally, instead of analyzing the entire genome, the method targets specific marker alleles. This approach maintains high accuracy while significantly reducing testing time and cost.
These specific marker alleles are known as “STRs” (Short Tandem Repeats). After amplification, they are compared to verify biological relationships such as paternity, parentage, or sibling relationships.
What Are STRs?
STRs (Short Tandem Repeats) are short, identical sequence units that are repeated consecutively within a DNA sequence.
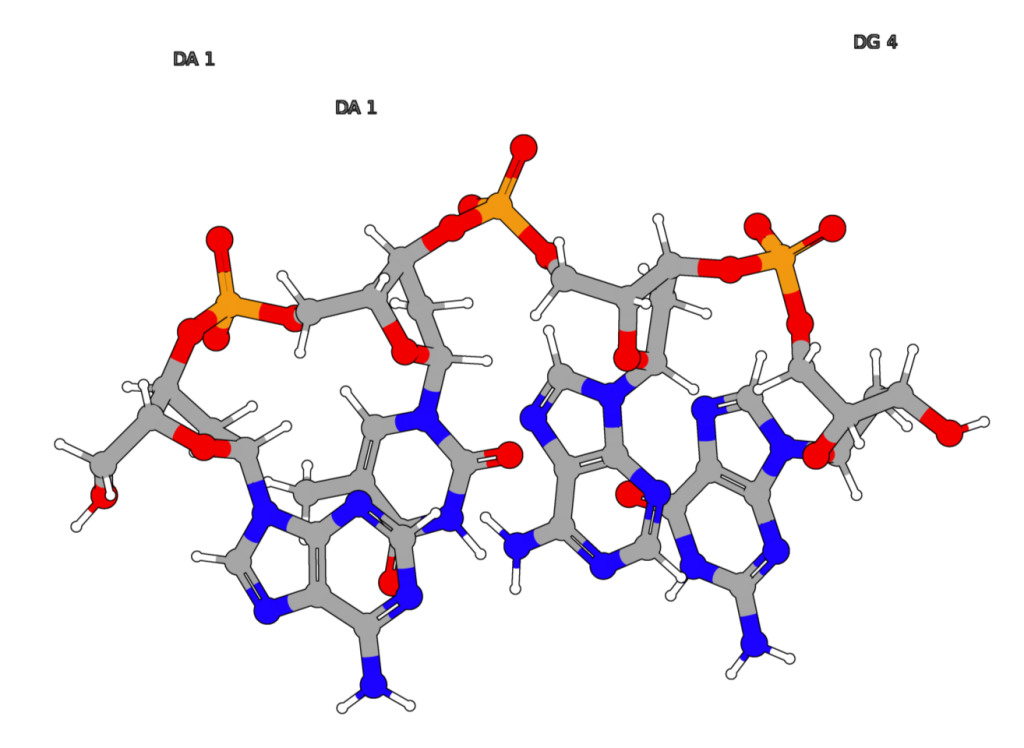
Figure 1: In the case of the STR known as CSF1PO, the short sequence “AGAT” is repeated between 5 and 17 times.
Figure 2: A schematic diagram showing three different STRs: D3S1358, D4S2408, and FGA.
There are various types of STRs depending on the specific sequence and location.
STR Analysis Method
Specific polymorphic short tandem repeat (STR) loci are first labeled through a two-step polymerase chain reaction (PCR) process, followed by targeted next-generation sequencing (NGS). The high read depth and sensitivity of this method allow for detection even from samples such as used toothbrushes or drinking straws.
The amelogenin gene is widely used for sex determination in forensic and prenatal diagnostics. In our testing, we enhance the accuracy and reliability of sex determination by analyzing an additional 25 Y-chromosome markers.
We employ the MiSeq FGx Sequencing System, the first next-generation sequencing platform specifically designed, developed, and validated for forensic genomics. After sequencing with this robust genetic panel, an integrated optimized analysis platform—the Universal Analysis Software (UAS)—performs bioinformatics analysis.This platform uses a proprietary alignment algorithm to check not only the STR repeat regions but also the flanking regions adjacent to the primer sequences (both forward and reverse). It identifies loci, alleles, and genotypes based on the entire amplicon sequence, rather than just amplicon size.
Loci with read counts below a specified threshold are excluded, while those above the threshold are considered valid alleles. Furthermore, the sequencing data is rigorously analyzed, taking into account important factors such as allele frequencies in populations, linkage disequilibrium, and the physical proximity of STR loci on the same chromosome.This results in highly sensitive, accurate, and reliable outcomes.
The systems and reagent kits used in this testing have undergone developmental validation and meet the minimum standards set by European guidelines, SWGDAM (Scientific Working Group on DNA Analysis Methods), CODIS (Combined DNA Index System), Interpol, and ESS (European Standard Set).
Details of the bioinformatics analysis performed by the proprietary Universal Analysis Software (UAS) and the statistical algorithms used for kinship analysis are beyond the scope of this article.
If you are interested in the mathematical foundations, please refer to the cited references for an overview of the methods employed.
- Kane K. (1982). Paternity exclusion and probability of paternity. Annals of clinical and laboratory science, 12(4), 309–314.
- Bugert, P., Rink, G., Kemp, K., & Klüter, H. (2012). Blood Group ABO Genotyping in Paternity Testing. Transfusion medicine and hemotherapy : offizielles Organ der Deutschen Gesellschaft fur Transfusionsmedizin und Immunhamatologie, 39(3), 182–186. https://doi.org/10.1159/000339235
- S. Arnott, P.J. Campbell-Smith & R. Chandrasekaran. Handbook of Biochemistry and Molecular Biology, 3rd ed. Nucleic Acids–Volume II, G.P. Fasman, Ed. Cleveland: CRC Press, (1976). pp. 411-422.
- Schrödinger, L., & DeLano, W. (2020). The PyMOL Molecular Graphics System, Version 3.1.0a0. Schrödinger, LLC.. Retrieved from http://www.pymol.org/pymol
- Tomasello, Gianluca, et al. ‘The Protein Imager: A Full-Featured Online Molecular Viewer Interface with Server-Side HQ-Rendering Capabilities’. Bioinformatics, edited by Arne Elofsson, vol. 36, no. 9, May 2020, pp. 2909–11. DOI.org (Crossref), https://doi.org/10.1093/bioinformatics/btaa009.
- Stephens, Kathryn M., et al. ‘Developmental Validation of the ForenSeq MainstAY Kit, MiSeq FGx Sequencing System and ForenSeq Universal Analysis Software’. Forensic Science International: Genetics, vol. 64, May 2023, p. 102851. DOI.org (Crossref), https://doi.org/10.1016/j.fsigen.2023.102851.
- Thangaraj, K., Reddy, A. G., & Singh, L. (2002). Is the amelogenin gene reliable for gender identification in forensic casework and prenatal diagnosis?. International journal of legal medicine, 116(2), 121–123. https://doi.org/10.1007/s00414-001-0262-y
- Jäger, AC, et al. Developmental validation of the MiSeq FGx Forensic Genomics System for targeted next generation sequencing in forensic DNA casework and database laboratories. Forensic Sci Int Genet. 2017; 28: 52–70. doi.org/10.1016/j.fsigen.2017.01.011.
- Nakazato T, Ohta T, Bono H. Experimental design-based functional mining and characterization of high-throughput sequencing data in the sequence read archive. PLoS One. 2013; 8 (10): e77910. doi.org/10.1371/journal.pone.0077910.
- Ballard D, Winkler-Galicki J, Wesoły J. Massive parallel sequencing in forensics: advantages, issues, technicalities, and prospects. Int J Legal Med. 2020; 134: 1291–1303.doi.org/10.1007/s00414-020-02294-0.
- Ingold S, et al. Body fluid identification using a targeted mRNA massively parallel sequencing approach – results of a EUROFORGEN/EDNAP collaborative exercise. Forensic Sci Int Genet. 2018; 34: 105–115. doi.org/10.1016/j.fsigen.2018.01.002.
- Bentley, DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008; 456: 53–59. doi.org/10.1038/nature07517.
- Huston, K. A. (1998). Statistical analysis of STR data. Profiles DNA, 1(3), 14-15.
- Evett, Ian, and B. S. Weir. Interpreting DNA Evidence: Statistical Genetics for Forensic Scientists. Sinauer Associates, 1998.
- Gjertson, D. W., Brenner, C. H., Baur, M. P., Carracedo, A., Guidet, F., Luque, J. A., … Morling, N. (2007). ISFG: Recommendations on biostatistics in paternity testing. Forensic Science International: Genetics, 1(3-4), 223–231. doi:10.1016/j.fsigen.2007.06.006






