Regarding the Test for 228 Types of Recessive Genetic Disorders
At Hiro Clinic, we offer a test that can examine 228 genes associated with severe recessive genetic disorders before birth.
Recessive genetic disorders are conditions that occur when there is an abnormality in the genes.
In this test, to determine whether both parents carry gene abnormalities, genes are extracted and analyzed from the oral mucosa of the mother and father.
By testing whether both parents have abnormalities in the same gene location, it is possible to predict the risk of the fetus having a severe genetic disorder.
For example, if both parents have abnormalities in the same gene, the child is at higher risk of developing a recessive genetic disorder (about 25%). If the test result is positive, amniocentesis is necessary as a confirmatory test.
So, what kind of test is conducted for such individuals? Also, what kind of disorders are identified?


It is recommended that couples/couples consider genetic testing
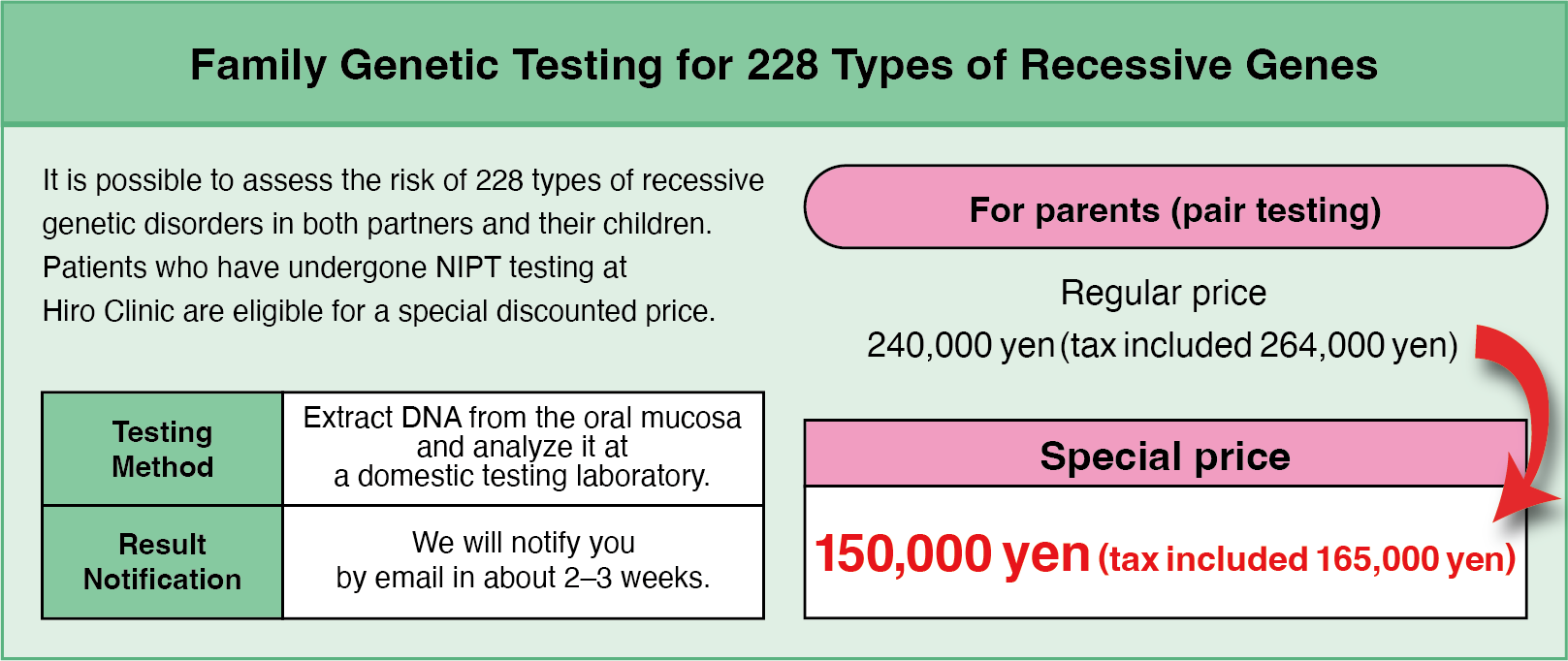
Hiro Clinic recommends that you undergo 228 different recessive gene tests. This allows you to find out about conditions that may be inherited by your child before birth. Knowing this information can be useful for future health and family planning, enabling you to plan early measures and support.
Benefits
Comprehensive risk assessment
The genetic information of the couple or couples can be used to extensively assess and take necessary action for diseases that may be inherited by their children.
Family planning support
Support decision-making to take genetic risk into account in future family planning.
Optimising preventive measures
Optimising preventive measures: individualised preventive measures and early intervention can be planned based on identified genetic risks.


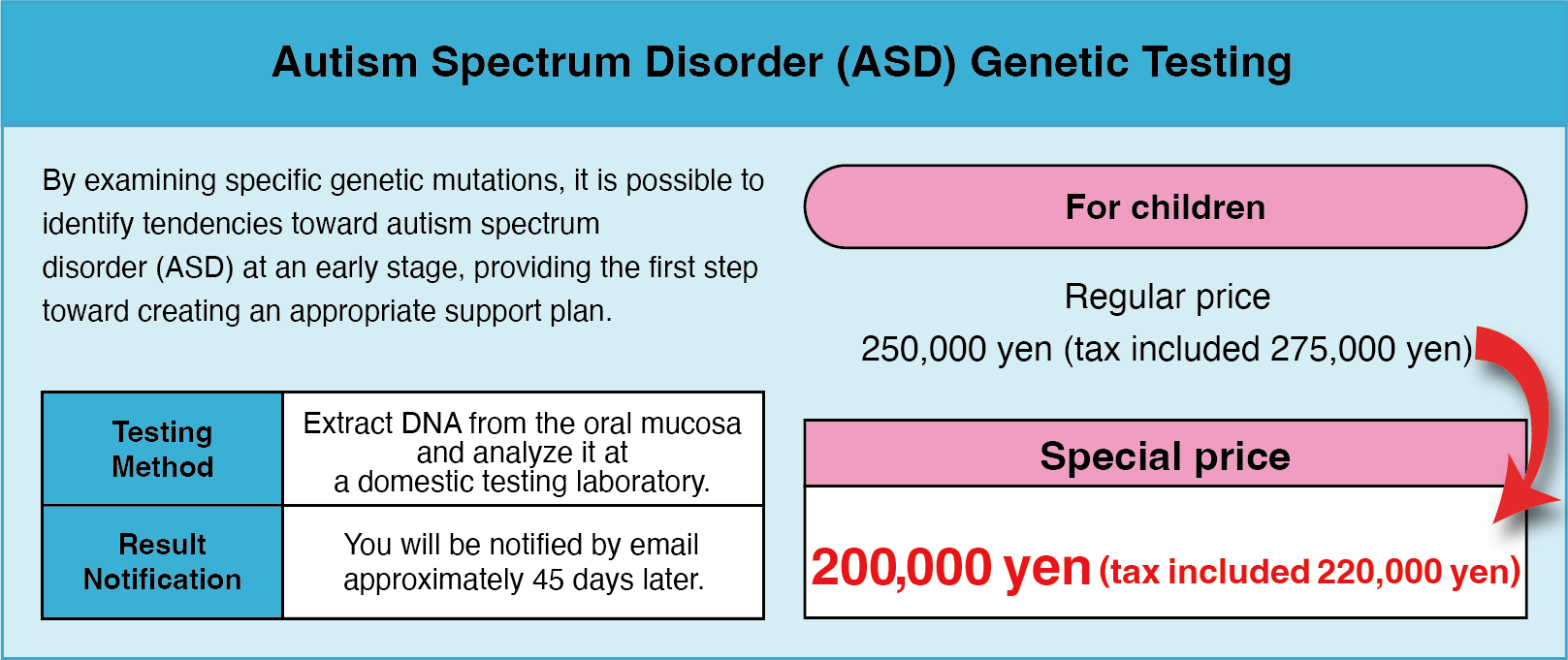
We recommend that you consider taking a genetic test for your newborn baby
Having your newborn baby undergo recessive genetic testing is one way to support your child’s health and future well-being. The test allows for early identification of genetic risks and, if necessary, appropriate preventive measures and treatment can be initiated immediately. At Hiro Clinic, we prioritise your child’s safety and comfort and use state-of-the-art technology to offer a painless ‘just rub the stick of the kit on the buccal mucosa (cheek in the mouth)’ test.
Benefits
Early detection
Screening for 228 recessive genetic conditions to detect potential risks to your child’s health at an early stage.
Individualised health care
Based on the test results, specific health care and treatment needs can be planned for your child.
Family security
Clear information about your child’s health status can help ease family anxiety.
Special limited price for those who have undergone Hiro Clinic NIPT
Hiro Clinic offers 228 recessive genetic tests at a special price to those who have undergone NIPT.
More information
If you have not undergone NIPT, don’t worry
228 recessive genetic tests are also available for those who have not undergone NIPT at Hiro Clinic.
More information
Inspection process
Optional Fees for 228 Types of Recessive Gene Tests
Available
How recessive genetic disorders develop
Humans have two chromosomes, one of maternal origin and one of paternal origin.
A recessive genetic disorder is a disease that develops when two chromosomes are abnormal in the same place. If one of the chromosomes of maternal or paternal origin is abnormal, but the other chromosome is not abnormal, the disease does not occur. In that case, the individual is said to be a carrier of the relevant genetic disorder.
Carriers are the ones who have been told since ancient times that they should not have children by blood.
For example, if there is a rare disease that affects only one in 40 000 people, it is estimated that there is one carrier in every 100 people.
This is because the calculation is ‘1 in 100 x 1 in 100 x 1 in 4 = 1 in 40,000’.
If we test 100 places for one disease in 40,000 people, this means that theoretically everyone could be a carrier of one disease.
So what happens if our test shows an abnormality in the same place in both parents’ genes?
Answer is
- One in four foetuses will develop the disease.
- One in two people will be a carrier.
- One in four individuals will be normal.
How can it be found out?
At present, the most accurate one is the amniotic fluid test.
This amniotic fluid test requires genetic analysis. Tokyo Hygienic Laboratory carries out this analysis in collaboration with Medicover in Cyprus.
If you have amniotic fluid testing support, the test usually costs less than 200 000 yen, so there is no cost burden.
Please contact Hiro Clinic for more information on amniotic fluid testing.
The 228 recessive genetic disorders are the following diseases.
Hiro Clinic NIPT can tell you about
Recessive genetic disorders
What are recessive genetic disorders at Hiro Clinic?
Hiro Clinic can detect 228 serious recessive genetic disease gene associations before birth.
This involves extracting and analysing genes from the buccal mucosa of the mother and father and combining them to detect serious genetic disorders in the foetus.
Serious genetic disorders are very rare, but even for a recessive genetic disorder that affects 1 in 40 000 people, the number of carriers of the disease (individuals with an abnormality in one of the two chromosomes but without the disease) is 1 in 100.
The HIRO Clinic considers the test to be useful in identifying carrier genes and suspecting a link between 228 different recessive genetic disorders based on their combination.
The unique feature of this test is that it detects all the combinations of genes that have been previously considered to be genetic abnormalities and determines whether the genetic changes are indeed pathogenic.
Genes differ from individual to individual. This is often the case for important genes, and their diversity makes each person different. However, they can also be pathogenic in their variation.
Gene combinations are shared in databases around the world, which are searched to see if any of the 228 gene variants are pathogenic.
When the mother and father are pathogenic at the same site, there is a high frequency risk of the child developing the disease. However, it is not enough just to look at the genetic change. It is necessary to determine (annotate) whether the genetic change is really abnormal. This is a joint effort between the Tokyo Health Laboratory and Medicover.
Testing for recessive genetic disease genes
228 carrier screening tests developed by the Medicover Genetics Ltd laboratory.
Genomic deoxyribonucleic acid (gDNA) is extracted using standardised methods and mechanically fragmented prior to DNA library preparation.
DNA enrichment of the genomic region of interest is performed using solution-based hybridisation methods and sequenced next-generation sequencing (NGS).
The read-out sequence data is referenced to a reference genome and mutations are identified using a proprietary bioinformatics pipeline.
Recessive inheritance can be identified for single nucleotide variants, small insertions and deletions (≤30 bases) and copy number variations (CNVs).
Variants are classified according to the American College of Medical Genomics and Genomics 3-5 criteria.
Variant classification and interpretation is performed using the Varsome Clinical platform and is based on information available at the time of testing.
Only pathogenic and suspected pathogenic mutations are reported. Variants of detected but unknown significance, benign or potentially benign mutations are not reported.
Genetic counselling is recommended for clinical interpretation and results.
A: If autosomal results are:
A-1: “No clinically significant variants detected”
While not a complete guarantee that the testee is not a carrier of a genetic disease, they indicate the absence of a genetic variant and are unlikely to be a carrier.
A-2: ‘Clinically significant variant detected’
Indicates that a genetic alteration has been identified and that the testee is a carrier of the disease.
The person can then be a carrier of two or more diseases.
Carriers usually do not have symptoms of the disease.
However, two genes on two chromosomes may both be abnormal, in which case the possibility that the testee is currently affected or will be in the future cannot be ruled out.
B: For X-linked inherited diseases:
B-1: ‘No clinically significant variant detected’
Indicates the absence of a heritable variant, and can be said to indicate that the testee is not affected if the testee is male, or is unlikely to be a carrier if the testee is female, but it cannot be ruled out completely.
B-2: ‘Clinically significant variant detected’:
Indicates that a genetic alteration has been identified. In the case of female test subjects, it is possible that they are carriers.
If the testee is male, it indicates that he is currently affected or may develop the disease in the future.
However, the disease groups in this panel vary in severity and may not present clinically.
The aim of the test is to detect all variants associated with the detected gene by targeting all coding exons, MANE and/or standard transcripts and 10 bp of adjacent intronic sequences.
Mutants outside the target region are not intended to be detected by this assay.
Unless otherwise stated, sequence changes (SNVs and INDELS) in promoters and other non-coding regions are not detected by this assay.
Specific sequence changes (SNVs and INDELS) in non-coding regions considered clinically important for the detected gene are included in the analysis.
If two variants are identified in a gene, it is not possible to distinguish whether these are on one chromosome (cis) or another (trans).
Genetic changes such as inversion, rearrangements, ploidy and epigenetic effects are not covered by this test.
Certain sequence variations in target regions, including repetitive sequences (SNVs and INDELS), highly homologous sequences such as segmental duplications and pseudogenes, and regions of high/low GC content may not be detected.
Copy Number Variations (CNVs) are calculated using high-quality sequencing reads that are free of duplicates and uniquely aligned.
CNVs are detected for a subset of target regions using GC content normalisation and depth of sequencing coverage approach.
CNV anomalies are detected if the detected coverage deviates significantly from the coverage estimated from the reference site.
CNVs can be detected down to a resolution of a few exon levels.
If the CNV is positive, it is confirmed using the orthogonal method.
CNVs cannot be detected in genomic regions that contain little or no mapping, repetitive sequences, pseudogenes or high/low GC content.
Detecting CNVs using NGS is less sensitive/specific than orthogonal quantification methods, so the absence of a reported CNV does not guarantee that it is not present.
The fact that no disease-causing variants are present in a target gene reduces the likelihood of disease, but does not completely exclude the possibility of disease-related syndromes.
Validation testing is carried out by Medicaver Genetics Ltd.
The test does not identify all mutations associated with the disease tested.
While the test is highly accurate, the possibility of false positives or false negatives still exists and can be caused by technical or biological limitations.
These include rare genetic variants, mosaicism, blood transfusions, bone marrow transplants or other rare molecular events.
Some undetected genetic alterations may be affected and are not tested by carrier screening tests.
Although genetic testing is an important part of the diagnostic process, genetic testing does not always give definitive answers. In some cases, a genetic mutation may be present but not identified by testing.
This is due to limitations in current medical knowledge or testing techniques.
Concurrent use of other clinical data and clinical findings is recommended.
Resulting results should always be considered in relation to other clinical findings.
The clinician who referred the patient for testing is responsible for pre- and post-test counselling, including advice on the need for additional genetic testing.
Other diagnostic tests may be required.
 中文
中文